Research
Molecular Signaling Networks that Govern (Myo)fibroblast Plasticity
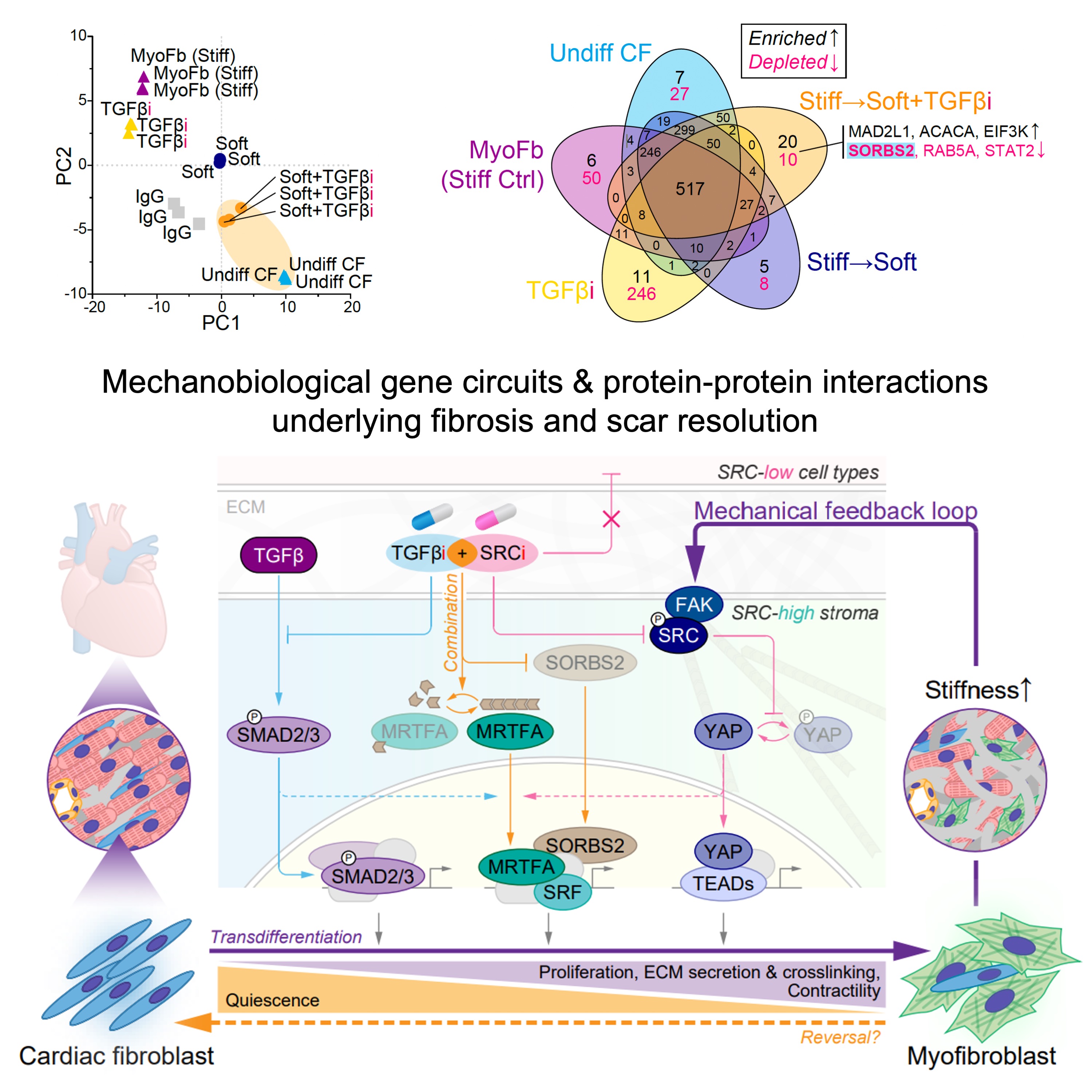
Our cells are remarkably responsive to a wide range of microenvironment cues—including both biochemical (soluble) signals and mechanical (insoluble) stimuli—but how these combined inputs are processed simultaneously remains incompletely understood. In the Cho Lab, we study under what circumstances soluble and insoluble cues synergize with or antagonize each other, and how this crosstalk influences: (i) cell state plasticity, (ii) sex- and disease-specificity, and (iii) intercellular communication mechanisms in fibrotic remodeling and tissue repair.
Cardiovascular Engineering Across Multiple Scales
Our team has extensive expertise in human induced pluripotent stem cell (iPSC)-based engineered tissues as well as several animal models. We routinely work with: (i) cells cultured on tunable biomaterials, (ii) arrays of multi-cell type cardiac organoids and spheroids, (iii) engineered heart tissue and 'heart-on-a-chip' systems, (iv) ex vivo chick embryonic hearts, and (v) mouse disease models. For every biological phemonon we study, we take a systems approach to examine, engineer, characterize, manipulate, and cross-validate across multiple scales.
Bioinformatics/AI-guided Precision Medicine
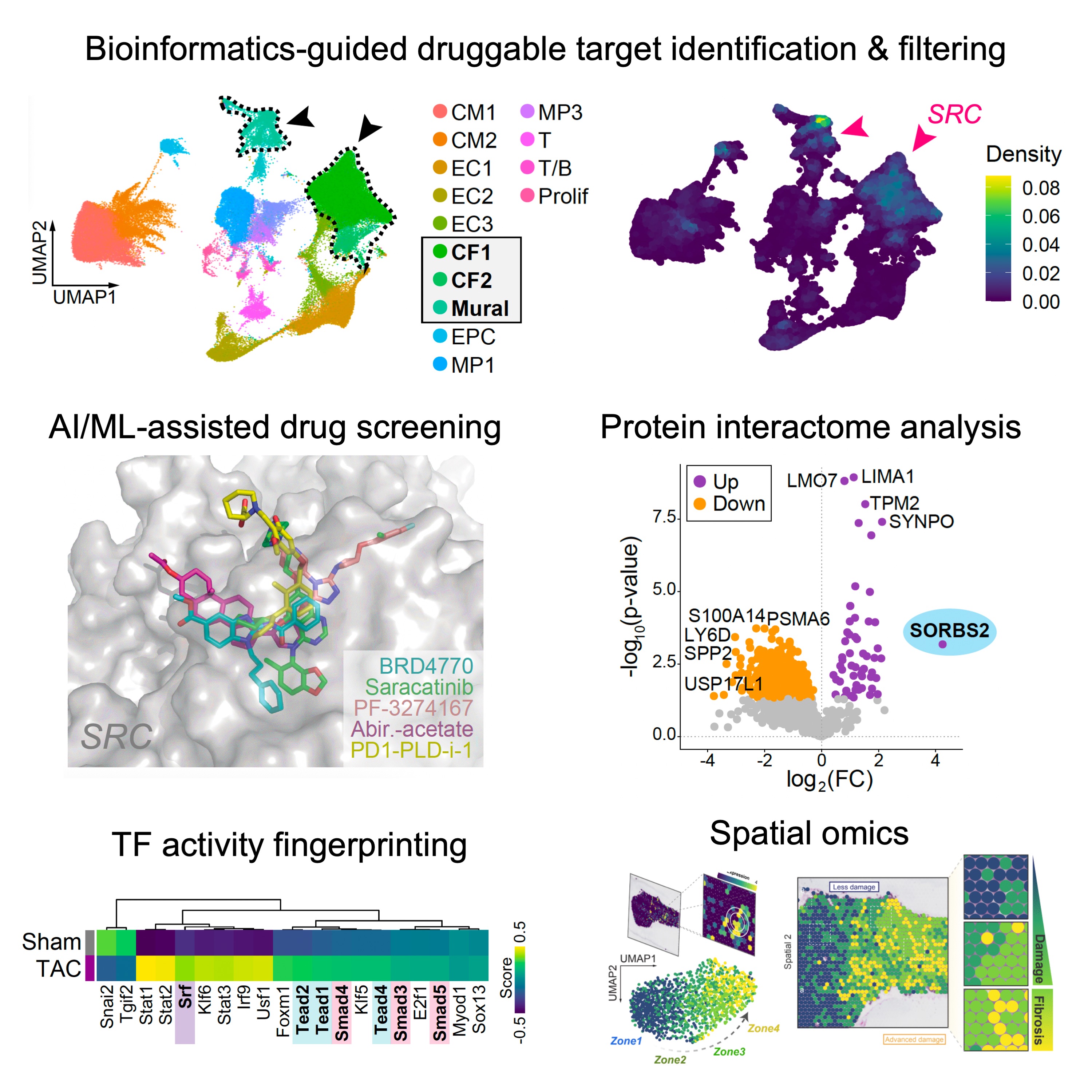
We combine molecular tools with cutting-edge multi-omics (scRNA- & scATAC-seq, spatial, and proteomics) and AI/ML-based methods to enable new druggable target identification, conduct in silico drug screening, map protein interactomes, and perform transcription factor activity fingerprinting. We are particularly interested in applying such tools to better characterize the functional diversity and state plasticity of cardiac stomal populations across various disesase states—a critical challenge for the development of precision anti-fibrotic therapies.
Recorded Talks
Check out our recent presentation at the Bay Area Cardiovascular Symposium